NA-sampler
This page describes the package NA-sampler. It consists of a set of user callable subroutines which implement theNA multi-dimensional parameter space search algorithm. The package contains source code for the NA routines and an example driver program. The latter applies the NA algorithm to the estimation of crustal seismic velocity profiles from inversion ofseismic receiver functions (RFI). Several independent `utility' programs are alsoprovided which manipulate the output files of the main program and plot some results. Some of these programs are specific to the RFI problem, and others may be more generally useful.
These pages are meant only as a guide. For more information look at the various README files in the src sub-directories, the comments in the source code for the RFI example, and the input files for the utility plot programs. Finally if you are that adventurous you can look at the code itself.
This package performs a derivative free parameter space search using the Neighourhood Algorithm. See references below for details. The latest details of NA-sampler and the related ensemble inference algorithm NA-Bayes can be found on the iEarth NA page
- Installation
- Using the NA subroutines
- Controlling the Neighbourhood Algorithm
- Tuning the control parameters
- Utility programs
- Running the RFI examples
- Conditions of use
- Contacting the author
- Papers and related www sites
Installation
When the tar file is unpacked it produces four directories:
src - contains all Fortran and C source code for NA subroutines and RFI example program and utility/plot programs.
data - contains data input files and example output files created by RFI example problem. Also contains data files for running splot graphics program, and RFI plot programs. (See utility programs for more details.)
bin - contains executable for RFI example program and all utility programs.
docs - contains these HTML pages.
To remove all object and source files prior to a re-installation type `make clean'. Examine the Makefile if you do not want to compile all of the utility programs. Note that the file `makena.macros' contains the location of the Fortran and C compilers and must be edited to suit your system prior to installation.Typical configuration files are provided for compaq alpha, sun, GNU, and Linux. These can be used by typing,
-
> cd src
> cp macros/makena.macros_sun makena.macros
To compile the complete set of programs from the src directory
-
> make all
If compilation is successful, all executables are placed in the bin directory and all examples demonstrating the utility programs can be run from the data directory (See below). The reverse of make all is,
-
> make clean
which will remove all .o files and all executables.
Note that the Demo directory, the Makefile and the file rfi_na.f can serve as a template for new user applications. If you are not interested in running, or looking at output from the seismic example program then you can remove all RFI specific source code and programs by,
-
> cd src
> \rm -fr rfi_subs
> cd ../data
> \rm -fr demo_results rfi_files
This will save disk space but please note that the RFI programs will be gone and the command `make all' will no longer work. A re-installation from a tar file will be necessary to retrieve the RFI example files.
Parallelization with MPI
In May 2002 message passing interface (MPI) calls were added to the NA source code. This allows the possibility of running the search algorithm across a cluster of machines simultaneously (e.g. a linux Beowulf cluster). The advantage of this is that the forward modelling for each model in an ensemble can be performed on separate machines, giving much faster overall run times. Note that this does not mean that the user's own forward modelling code needs to be parallelized with MPI. All MPI calls appear in the NA source code and are used to farm the forward modelling out to different processors.
The MPI code is activated by a switch in the installation file `makena.macros'. If you have say a linux cluster running MPI then you need to set the TYPE=mpi and the MF77 variable to the location of the MPI fortran 77 compiler on your system. See the example makena.macros files for examples. (If you want the usual single processor version then set TYPE=serial.) If installation occurs with these variables then you must execute the program using the mpirun command provided by your MPI installation in the usual way. For example one might run the seismic example executable with a command like,
-
> mpirun -np 20 rfi_na
which will execute the program rfi_na with the forward modelling distributed across the first 20 machines that your MPI is set up to run on.
Note that when the number of processors (e.g. 20 in this example) divides equally into the number of models generated at each iteration (see parameter Ns2 below) then one gets perfect parallelization in the forward modelling part of the code, and this usually means a significant overall speed up, e.g. a factor of Ns1/20 in the forward modelling part of the code. Since for many applications most time will be spent on forward modelling then this can represent a significant speed up of the overall code. When the number of processes is not a factor of Ns2 then one gets a smaller speed up factor. Note that there is no point in asking for more processors than Ns2 because there will be nothing for the extra processors to do.
Compiling problems and platform dependency
So far I have succesfully compiled and run all codes in this package on a SUN workstation (running Solaris 2.6 or later) using both the native compiler and the GNU compilers g77 and gcc; on a Compaq Alpha (running OSF1 V4.0) using the Digital Fortran and C compilers; on a linux box with g77, or intel fortran/C compilers ifc and icc. In all cases exactly the same results were obtained. Using the intel fortran and C compilers on Linux I obtained different answers. Possibly due to rounding error changing the Monte Carlo sequence. The default makena.macros file is for a linux box running the intel compiler. When installing on other platforms SUN you will need to edit the file src/makena.macros appropriately.
If you are using a Dec/Compaq alpha/compaq platform then you will need to use the compiling option -assume byterecl which makes the Fortran open statement for direct access files the same as on a sun, i.e. with record length specified in bytes rather than words. This can be done by editing the file src/makena.macros, which is used to define compilers and options during the make. An example file makena.macros_alpha is provided as an example.
Note that for the example problem direct access output (nad) files are provided with the package (in directory data/demo_results). To allow platform independence both little endian and big endian binary versions are included. The make file uses your choice of platform (specified in file makena.macros) to decide which type of binary file to link to the examples. This is a change over earlier distributions and means that you will now be able to run all of the utility programs, e.g. splot (showing the models generated by the NA ) on the example output files provided. The downside of this is that the distribution tar file is larger.
Some programs (from T. Shibutani, RCEP, DPRI, Kyoto Univ., Japan) are provided in the directory src/utl/byteconv for converting SAC files between a sun and a Compaq Alpha machine. This may be useful for seismologists with many sac files. Note that SAC files are the format used to store the input and output seismic receiver functions used in the RFI example. They are specific to this example and are read in by the user routines. They are not a generic NA input file.
Fortran 90
In January 2004 the source codes of the NA_sampler package were changed to incorporate dynamic memory allocation using Fortran 90. This means that array bounds in file na_param.inc only need to be set if using Fortran 77 compatibility mode, which is itself set by -DNA_F77_COMPATIBILITY=1 in the file makena.macros. Try the following
-
> cd src/macros
> diff makena.macros_alpha_f90 makena.macros_alpha_f77
Fortran 90 will be the default compiler from January 2004.
Code structure and interfacing
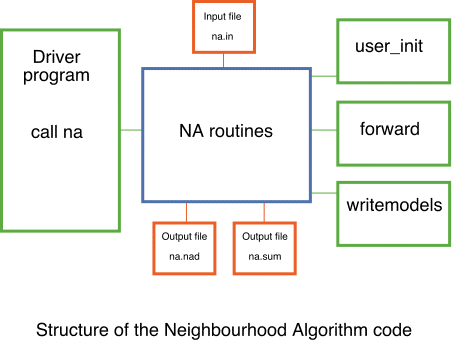
The figure above shows the structure of the NA code and how it is interfaced with the user code. The green routines must be provided by the user. The main routine passes control to the NA code by calling the subroutine `na' with no arguments. This then calls three user written subroutines.
user_init - provides the definition of the parameter space to the NA routines and performs any user problem specific tasks and I/O.
forward - calculates the misfit/objective function for a given input model.
writemodels - allows the user to write out, in their own format, all of the models, misfit values etc, from the ensemble produced by the NA.
Only forward is called more than once. Each of these subroutines are fully described in the header of the example source files for the RFI problem.
The file rfi_na.f contains the main driver program and the three subroutines which implement the RFI example problem. In this case the driver routine merely calls na and exits. All data I/O and initialization is performed inside the routine user_init. The three RFI interface routines make use of many other problem specific subroutines and libraries. Source files for all of these are in the directory src/rfi_files. They are compiled and linked into the main program by the Makefile.
To use the NA subroutines you need to do the following,
-
Create the three subroutines for your problem. (Use the RFI examples as a template)
-
Create the main routine that calls na (This can be trivially simple as in the RFI example.)
-
Edit the file na_param.inc to adjust array sizes if necessary. Since January 2004 this is not needed if using Dynamic memory allocation under the fortran 90 compiler (set NA_F77_COMPATIBILITY=0 in makena.macros)
-
Edit the Makefile and makena.macros to compile the users subroutines
-
Compile the new program with
- > make rfi_na (or the equivalent)
Note that the only essential information that the NA routines need from the user routines is the definition of the parameter space (from user_init) and the misfit for a given model (from forward). The routine writemodels is called once after the NA has finished and can be null.
Output files
The two generic output files are na.sum and na.nad The first is an ascii file with summary information on the performance, the second is a direct access file binary file containing all of the models generated by the NA during the search. The format of this file is described in Appendix C. The NAD file is used as a compact storage format which allows users to add their own information in a header. NAD files are read in by certain utility programs like the plot program splot (See appendix A). NAD files are also read in by the appraisal algorithm NA-Bayes, which is a separate package that can be downloaded. See the NA homepage for details.
Code development
The development of this code is an ongoing research effort and I expect that the NA subroutines will be updated from time to time, however, it is intended that the interface routines will remain unchanged. (The input file na.in will probably change) In this way the incorporation of updated NA routines into an existing application should be quite straightforward, i.e. compiling the new na.f to obtain na.o and linking with the existing driver code. Note that the NA source code in sub-directory NA_src should not normally need editing ! (Edit at your own risk !)
The code was developed on a sun network under Solaris 2.6. Note that there are some issues to consider when transporting the code across to new platforms and compilers systems. The ones that I am aware of are described in the section on Compiling problems.
Controlling the Neighbourhood algorithm
The NA control parameters and other options are read in from the file
na.in.
Each line is described below.
-
#
# Neighbourhood Algorithm input options file
#
ialg : Algorithm type (1=Uniform search, 2=Neighbourhood algorithm)
maxit : Maximum number of iterations
Ns1 : Number of models for initial sample
Ns2 : Number of models generated at each iteration
Nr : Number of Neighbourhoods re-sampled at each iteration
YorN, iseed : Use quasi random number generator ? (y/n); random seed
istype : Type of initial sample (0=random;+-1=read in a NAD file)
olevel : Output info level (0=silent;1=summary info;2=1+write models)
YorN : Turn timing mode on ? (y/n)
YorN : Turn debug mode on ? (y/n)
Most of these are self-explanatory. The examples in the data directory contain `typical' values for the parameters, although the main tuning parameters Ns2 and Nr should be tuned for the particular problem. (See tuning the control parameters)
The parameter istype controls the type of initial ensemble of models used. Several options are available.
-
istype=0 - then the initial sample is generated using a uniform random distribution and has size Ns1.
-
istype=1 - then the initial sample is read in from a direct access file in nad format called `na.nad' and the initial sample size Ns1 is reset to the number of models in the nad file.
-
istype=-1 - same as istype=1 except that the forward problem is not solved for the initial ensemble and instead the misfit values are taken from the file.
-
istype=2 - then the initial sample is read in from an ascii file models.in. For each model the format is a triplet of a blank line followed by the misfit value on a line by itself followed by the nd values of the model. This is repeated for each model. See the data directory for an example. If the number of models in the file is less that Ns1 then the remaining models are generated randomly. If greater than Ns1, then Ns1 is reset to that value.
-
istype=-2 - same as istype=2 except that the forward problem is not solved for the initial ensemble and instead the misfit values are taken from the file.
With these options it is possible to restart the NA using the output ensemble from a previous run of NA (istype=1). This might be useful if one wanted to change the tuning parameters (Ns2 and Nr), or perhaps change the misfit function its self (say to add regularization or incorporate some new type of data), and continue on. If istype=+-2 then its possible to seed the initial distribution with one or more previously known `good' models with low misfit. This will naturally attract the sampling towards them and allow a gradually focusing search in their neighbourhood. Examples of programs which read and write NAD files are contained in the utility programs section.
If a quasi random number generator is not chosen then a pseudo-random number generator is used. (See the papers cited below or Press et al 1992 Numerical Recipes, C.U.P., for a discussion of the difference.)
Tuning the control parameters
The two control parameters usually need to be tuned for the problem in hand and the style of sampling that you want. There are no well defined rules but experience has proven to be a useful guide. Most of my understanding is based on empirical evidence from applications at RSES. The usual objective with a direct sampling algorithm is to try and achieve a balance between exploration of the parameter space and exploitation of the previous samples. The more explorative an algorithm is, the more likely it will be robust against local minima, and the starting random seeds, the more exploitative it is the faster it will reduce the objective function. The amount of exploration and exploitation performed by the NA is largely controlled by the two control parameters ns (ns2) and nr.
- ns = Number of models generated at each iteration.
nr = Number of Voronoi cells re-sampled at each iteration.
- and so ns/nr models are placed in each cell.
See the papers below for more details. As ns and nr are increased it will be more explorative and as they are decreased it will be more exploitative.
Our experience has shown that reasonable results can be obtained after enough iterations when ns is of the order of twice the number of dimensions, and nr is in the range 2 - ns/2. The number of iterations is more difficult to judge. I have used used 10 - 100 times ns. It is clear that the larger values of ns and nr (i.e. more explorative) require more iterations to concentrate sampling (especially in high dimensions). In our experiments we have found in all cases that the sampling does eventually concentrate quite well, even for larger nr values, although you might not want to wait that long!
My strategy is often that it is better to get good sampling slowly than poor sampling quickly. However, small values of nr do seem to concentrate sampling quickly, especially in small dimensional (e.g. d ~ 4) problems. These issues are also discussed in Sambridge (1999a). I would be interested to hear of other peoples experiences (see contacting the author ).
Utility programs
When the package is compiled the following general utility programs are put in the bin directory (source in src/utl).
readnad - reads in a nad file and writes summary information to the screen.
- To read a nad file called `na.nad' then type
- readnad na.nad
nad2asc - reads in a nad file and writes out all models to an ascii file
- Execute with
- nad2asc na.nad outputfile
asc2nad - goes the other way
- Execute with
- asc2nad inputfile output.nad
splot - reads in a nad file and plots a scatter diagram of all models (PS or X)
- projected onto a pair of axes. See the
splot manual for more details.
The following are utility programs specific to the receiver function example:
plot_rf - plots the observed and predicted receiver function
-
for the best fitting velocity profile found by the NA.
plot_model - a density plot of all velocity profiles produced by the NA search.
synrf - calculates a synthetic receiver function from an input velocity profile.
synrf_noise - Same as synrf but adds noise to the result.
nad2rfi - converts direct access NAD file to an ascii file containing
-
velocity profiles, in a format specific to RFI problem.
rfi2nad - performs opposite conversion to nad2rfi.
Executables from all utility programs are put in the bin directory. Further details can be found at the RFI utility programs page.
Running the receiver function inversion example
The example driver program provided with the NA-sampler package is for the inversion of receiver functions for crustal seismic structure. This is described further in the two NA papers cited below. Note that the driver program is not intended to be a general purpose receiver function code, but merely an example of using the NA routines on a realistic seismic problem. Changes to the code may be required if it is applied to other data sets (For example, the maximum number of layers, and waveforms may need adjusting etc.)
To run the RFI examples after compilation of the package (see above),
-
> cd data
> cp na.in_examples/na.in_9_2 na.in
> rfi_na
This will run a short (3 iteration) test run with Ns=9 and Nr=2. The output should be similar to that in demo_results/na.sum_3_9_2. A similar set of steps is required to run one of the longer example inputs in directory na.in_examples. You could try editing na.in to adjust input parameters and repeating the test run.
The RFI subroutines use several input and output files. All of which are in sub-directories of data/rfi_files. The main one is rfi.in, which contains parameters associated with the input data and the names of the other I/O files.
Two other input files are used. In the default setup these are:
rfi_param - in sub-directory NA_MDL, which contains the bounds of the model parameters (Thicknesses, velocities and Vs/Vp ratio's in each of 6 layers).
rec_func - in sub-directory ORF, which contains the one or more input (stacked) receiver functions.
Two output files are produced. In the default setup these are:
rec_models - in sub-directory NA_MDL, which contains all models generated at each iteration of the NA in a convenient format.
rec_func - in sub-directory NA_SRF, which contains the predicted receiver function from the best fit model.
The output files produced by a 500 iteration example are included in the demo_results directory. They can be used to demonstrate the use of splot and the RFI graphics programs. See the splot manual for examples of using splot on the 500 iteration results. See RFI utility programs for examples of plotting receiver functions and the ensemble of velocity profiles produced by the NA.
Conditions of use
- The NAB routines may not be distributed to third parties.
Interested parties should contact the author directly. - Due acknowledgment must be made of the use of the NA routines in research reports or publications. Whenever such reports are released for public access, a copy should be forwarded to the author.
- The NA routines may only be used for not-for-profit research and development, unless it has been agreed in advance with the author in writing.
Use of any part of this package will be taken as acceptance of these conditions.
Contacting the author
The purpose of this documentation and the associated README files, is to reduce the likelihood of running into problems. I would be grateful if you do not contact me with queries on the installation, or running, of the code unless you have thoroughly looked for the answer yourself. (Note that either these HTML pages, the README files, example files and the source code itself may provide an answer). Note that for detailed queries on the RFI example it may be best to contact Dr. T. Shibutani, (shibutan@rcep.dpri.kyoto-u.ac.jp) who wrote the original versions of the RFI code and plot programs. His contribution is much appreciated.
I am interested in obtaining feedback on people's experiences with this code. Especially to hear what type of problems it has been applied to. I expect that the code will be updated from time to time. Information on new developments will be posted on the NA homepage. If you develop your own web pages showing work where NA has been used, or publish papers then I would be happy to add a reference on the NA homepage.
Related sites:
NA homepage - where the latest versions of NA-Bayes and NA-sampler can be accessed.
splot - graphics program for display of multi-dimensional ensembles.
NAD - explanation of direct access nad files.
RFI - Receiver function utility programs and graphics (included with NA-Sampler package.)
NA-Bayes - the manual for the neighbourhood algorithm Bayesian inference package.
naplot - graphics program for displaying Bayesian estimators (included with NA-Bayes package).
summary.pdf - A short summary of NA-sampler written for the IASPEI handbook (PDF file)
References:
The Neighbourhood algorithm for searching a multi-dimensional parameter space for models (points) with `acceptable data fit' is implemented in the program NA-sampler, and is described in the paper:
Geophysical Inversion with a Neighbourhood Algorithm -I. Searching a parameter space, Sambridge, M., Geophys. J. Int., 138 , 479-494, 1999. doi:10.1046/j.1365-246X.1999.00876.x
A related paper describes the Neighbourhood re-sampling algorithm (implemented in program NA-bayes) for calculating Bayesian integrals from a finite set of samples produced by the NA algorithm or any other search method (e.g. GA or SA etc.):
Geophysical Inversion with a Neighbourhood Algorithm -II. Appraising the ensemble,Sambridge, M., Geophys. J. Int., 138 ,727-746, 1999. doi:10.1046/j.1365-246x.1999.00900.x










